Synthesis and Conformational Analysis of Efrapeptins
09-Jan-2012
Chemistry, 2012, DOI: 10.1002/chem.201102134, Volume 18, Issue 2, pages 478–487 published on 09.01.2012
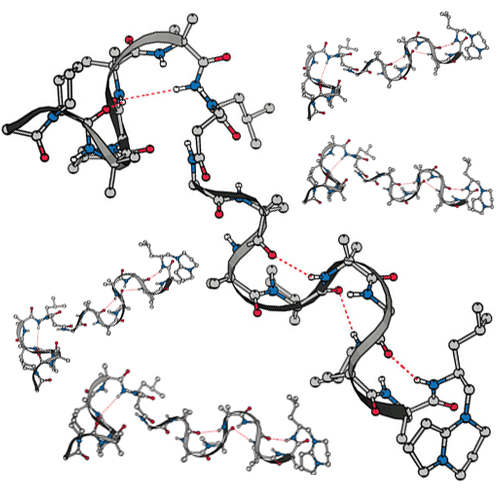
The efrapeptin family of peptide antibiotics produced by the fungus Tolypocladium niveum, and the neo-efrapeptins from the fungus Geotrichum candidumare inhibitors of F1-ATPase with promising antitumor, antimalaria, and insecticidal activity. They are rich in Cα-dialkyl amino acids (Aib, Iva, Acc) and contain one β-alanine and several pipecolic acid residues. The C-terminus bears an unusual heterocyclic cationic cap. The efrapeptins C–G and three analogues of efrapeptin C were synthesized using α-azido carboxylic acids as masked amino acid derivatives. All compounds display inhibitory activity toward F1-ATPase. The conformation in solution of the peptides was investigated with electronic CD spectroscopy, FT-IR spectroscopy, and VCD spectroscopy. All efrapeptins and most efrapeptin analogues were shown to adopt helical conformations in solution. In the case of efrapeptin C, VCD spectra proved that a 310-helix prevails. In addition, efrapeptin C was conformationally studied in detail with NMR and molecular modeling. Besides NOE distance restraints, residual dipolar couplings (RDC) observed upon partial alignment with stretched PDMS gels were used for the conformational analysis and confirmed the 310-helical conformation.