Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3
14-Jul-2015
PNAS, vol. 112 no. 28, 8614–8619, doi: 10.1073/pnas.1504648112
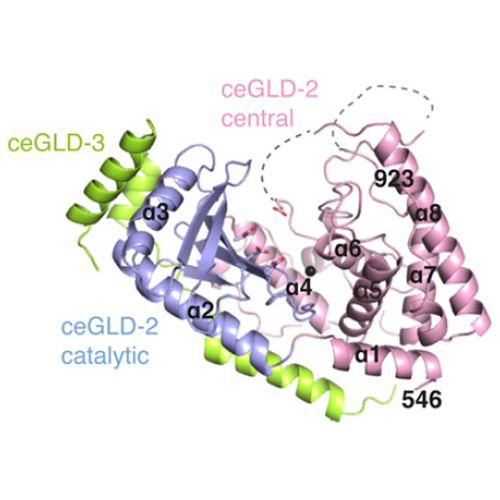
The Caenorhabditis elegans germ-line development defective (GLD)-2–GLD-3 complex up-regulates the expression of genes required for meiotic progression. GLD-2–GLD-3 acts by extending the short poly(A) tail of germ-line–specific mRNAs, switching them from a dormant state into a translationally active state. GLD-2 is a cytoplasmic noncanonical poly(A) polymerase that lacks the RNA-binding domain typical of the canonical nuclear poly(A)-polymerase Pap1. The activity of C. elegans GLD-2 in vivo and in vitro depends on its association with the multi-K homology (KH) domain-containing protein, GLD-3, a homolog of Bicaudal-C. We have identified a minimal polyadenylation complex that includes the conserved nucleotidyl-transferase core of GLD-2 and the N-terminal domain of GLD-3, and determined its structure at 2.3-Å resolution. The structure shows that the N-terminal domain of GLD-3 does not fold into the predicted KH domain but wraps around the catalytic domain of GLD-2. The picture that emerges from the structural and biochemical data are that GLD-3 activates GLD-2 both indirectly by stabilizing the enzyme and directly by contributing positively charged residues near the RNA-binding cleft. The RNA-binding cleft of GLD-2 has distinct structural features compared with the poly(A)-polymerases Pap1 and Trf4. Consistently, GLD-2 has distinct biochemical properties: It displays unusual specificity in vitro for single-stranded RNAs with at least one adenosine at the 3′ end. GLD-2 thus appears to have evolved specialized nucleotidyl-transferase properties that match the 3′ end features of dormant cytoplasmic mRNAs.