Structural basis for ATP-dependent chromatin remodelling by the INO80 complex
11-Apr-2018
Nature, volume 556, pages386–390 (2018), doi:10.1038/s41586-018-0029-y
Nature, online article
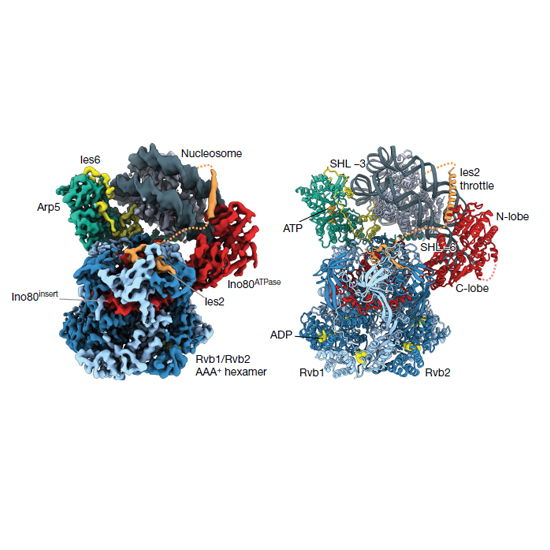
In the eukaryotic nucleus, DNA is packaged in the form of nucleosomes, each of which comprises about 147 base pairs of DNA wrapped around a histone protein octamer. The position and histone composition of nucleosomes is governed by ATP-dependent chromatin remodellers such as the 15-subunit INO80 complex. INO80 regulates gene expression, DNA repair and replication by sliding nucleosomes, the exchange of histone H2A.Z with H2A, and the positioning of + 1 and −1 nucleosomes at promoter DNA. The structures and mechanisms of these remodelling reactions are currently unknown. Here we report the cryo-electron microscopy structure of the evolutionarily conserved core of the INO80 complex from the fungus Chaetomium thermophilum bound to a nucleosome, at a global resolution of 4.3 Å and with major parts at 3.7 Å. The INO80 core cradles one entire gyre of the nucleosome through multivalent DNA and histone contacts. An Rvb1/Rvb2 AAA+ ATPase heterohexamer is an assembly scaffold for the complex and acts as a ‘stator’ for the motor and nucleosome-gripping subunits. The Swi2/Snf2 ATPase motor binds to nucleosomal DNA at superhelical location −6, unwraps approximately 15 base pairs, disrupts the H2A–DNA contacts and is poised to pump entry DNA into the nucleosome. Arp5 and Ies6 bind superhelical locations −2 and −3 to act as a counter grip for the motor, on the other side of the H2A–H2B dimer. The Arp5 insertion domain forms a grappler element that binds the nucleosome dyad, connects the Arp5 actin-fold and entry DNA over a distance of about 90 Å and packs against histone H2A–H2B near the ‘acidic patch’. Our structure together with biochemical data suggests a unified mechanism for nucleosome sliding and histone editing by INO80. The motor is part of a macromolecular ratchet, persistently pumping entry DNA across the H2A–H2B dimer against the Arp5 grip until a large nucleosome translocation step occurs. The transient exposure of H2A–H2B by motor activity as well as differential recognition of H2A.Z and H2A may regulate histone exchange.