Molecular architecture of the HerA–NurA DNA double-strand break resection complex
20-Dec-2014
FEBS Letters, 2014, DOI: http://dx.doi.org/10.1016/j.febslet.2014.10.035, Volume 588, Issue 24, Pages 4637–4644 published on 20.12.2014
FEBS Letters, online article
FEBS Letters, online article
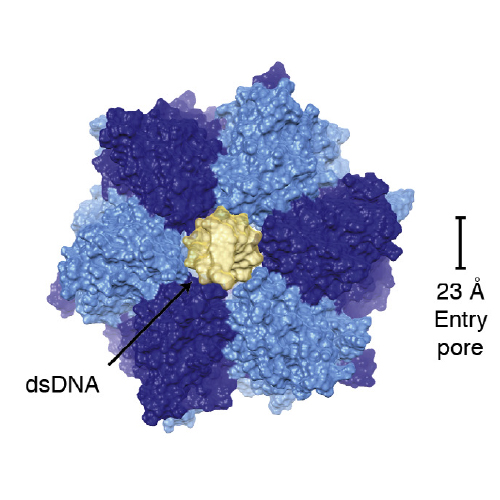
DNA double-strand breaks can be repaired by homologous recombination, during which the DNA ends are long-range resected by helicase–nuclease systems to generate 3′ single strand tails. In archaea, this requires the Mre11–Rad50 complex and the ATP-dependent helicase–nuclease complex HerA–NurA. We report the cryo-EM structure of Sulfolobus solfataricus HerA–NurA at 7.4 Å resolution and present the pseudo-atomic model of the complex. HerA forms an ASCE hexamer that tightly interacts with a NurA dimer, with each NurA protomer binding three adjacent HerA HAS domains. Entry to NurA’s nuclease active sites requires dsDNA to pass through a 23 Å wide channel in the HerA hexamer. The structure suggests that HerA is a dsDNA translocase that feeds DNA into the NurA nuclease sites.