Detection of Hydrogen Bonds in Dynamic Regions of RNA by NMR Spectroscopy
12-Dec-2014
Nucleic Acid Chemistry, 2014, DOI: 10.1002/0471142700.nc0722s59, 7.22.1-7.22.19, published on 12.12.2014
Nucleic Acid Chemistry, online article
Nucleic Acid Chemistry, online article
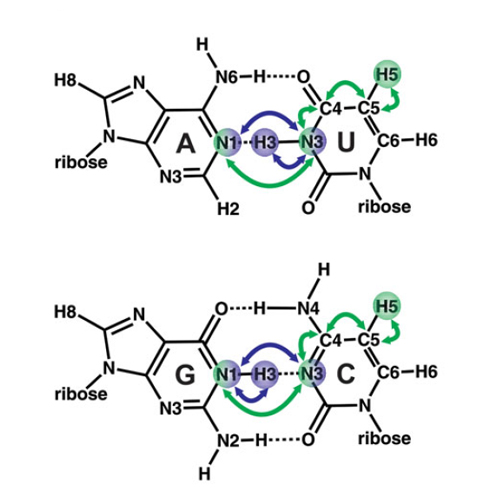
NMR spectroscopy is a powerful tool to study the structure and dynamics of nucleic acids. In this unit, we give an overview of important experiments to determine and characterize hydrogen bonds in nucleic acids and provide detailed instructions for setting up recently developed sensitivity-improved NMR pulse sequences, i.e., BEST selective long-range HNN-COSY, selective BEST-TROSY-HNNCOSY, and Py H(CC)NN-COSY. The strengths and limitations of these experiments will also be discussed. Detailed step-by-step protocols are provided for each of the three pulse sequences, with special emphasis on adjusting and setting of delays and shaped pulses. The NMR pulse sequences with example datasets and optimized, nonstandard adiabatic pulse shapes used for selective 15N magnetization transfer are provided. These experiments enable NMR analysis of a broad variety of RNAs ranging from low to high molecular weight and complexity.