Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution
16-Nov-2010
PNAS, 2010, doi: 10.1073/pnas.1009999107, vol. 107 no. 46 19748-19753 published on 16.11.2010
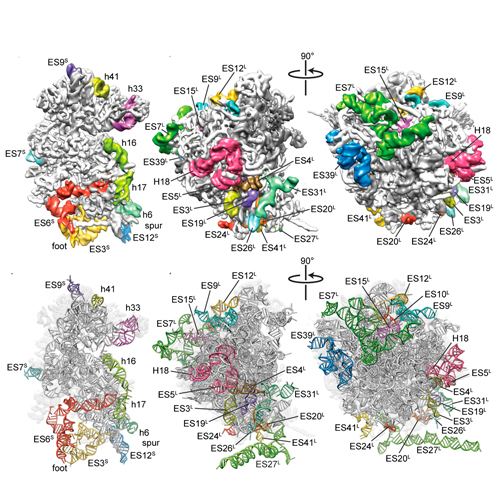
Protein biosynthesis, the translation of the genetic code into polypeptides, occurs on ribonucleoprotein particles called ribosomes. Although X-ray structures of bacterial ribosomes are available, high-resolution structures of eukaryotic 80S ribosomes are lacking. Using cryoelectron microscopy and single-particle reconstruction, we have determined the structure of a translating plant (Triticum aestivum) 80S ribosome at 5.5-Å resolution. This map, together with a 6.1-Å map of a Saccharomyces cerevisiae 80S ribosome, has enabled us to model ∼98% of the rRNA. Accurate assignment of the rRNA expansion segments (ES) and variable regions has revealed unique ES–ES and r-protein–ES interactions, providing insight into the structure and evolution of the eukaryotic ribosome.