Breaking the protein-RNA recognition code
31-Dec-2014
Cell Cycle, 2014, DOI:10.4161/15384101.2014.986625, Volume 13, Issue 23, published on 31.12.2014
Cell Cycle, online article
Cell Cycle, online article
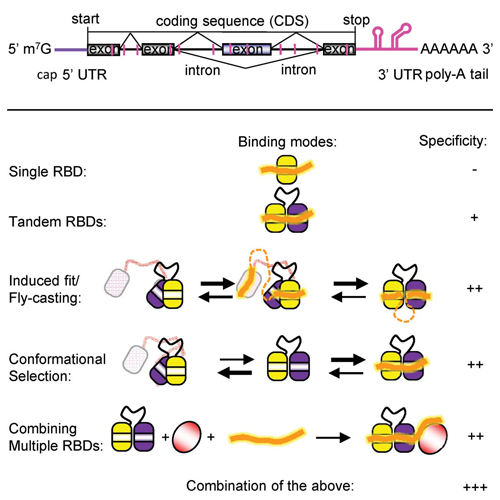
Recognition of mRNA transcripts by RNA binding proteins plays essential roles at various steps of gene expression, including splicing, translation, stability and general mRNA metabolism. Also the biogenesis and activity of non-coding RNAs (miRNAs, siRNAs, lncRNAs) and other regulatory RNA molecules depends on protein interactions. Cis elements, i.e. short RNA sequence motifs, in the mRNA are often degenerate and of low sequence complexity, for example, poly- (U) motifs that consist of a stretch of uridines. 1 Recent genome-wide studies start to provide a catalog of cis regulatory elements to which RNA binding proteins (RBPs) are bound.2 In parallel, the repertoire of trans-acting factors, i.e., RNA binding proteins is being established using high-throughput approaches.3 Although it has been found that many so far unknown RNA binding proteins (RBPs) may exist, most RNA interactions rely on a limited number of RNA binding domains (RBDs).3 It is still poorly understood how this limited set of RBDs can achieve sufficient specificity to unambiguously identify and bind to specific cis elements within the RNA and thereby regulate distinct processes.