Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P
02-Nov-2017
Molecular Cell, Volume 68, Issue 3, p515–527.e6, DOI: https://doi.org/10.1016/j.molcel.2017.10.014
Molecular Cell, online article
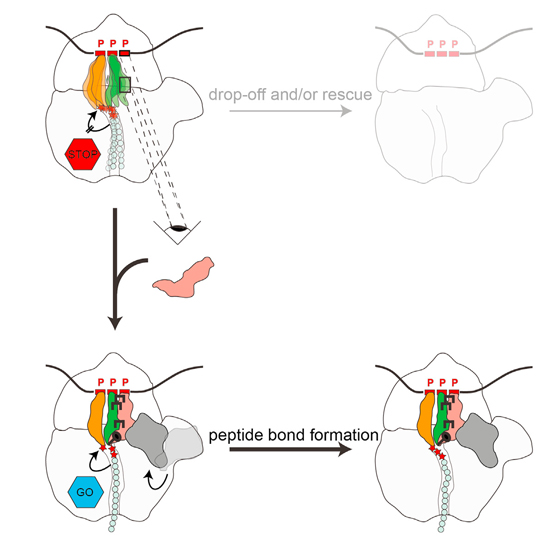
Ribosomes synthesizing proteins containing consecutive proline residues become stalled and require rescue via the action of uniquely modified translation elongation factors, EF-P in bacteria, or archaeal/eukaryotic a/eIF5A. To date, no structures exist of EF-P or eIF5A in complex with translating ribosomes stalled at polyproline stretches, and thus structural insight into how EF-P/eIF5A rescue these arrested ribosomes has been lacking. Here we present cryo-EM structures of ribosomes stalled on proline stretches, without and with modified EF-P. The structures suggest that the favored conformation of the polyproline-containing nascent chain is incompatible with the peptide exit tunnel of the ribosome and leads to destabilization of the peptidyl-tRNA. Binding of EF-P stabilizes the P-site tRNA, particularly via interactions between its modification and the CCA end, thereby enforcing an alternative conformation of the polyproline-containing nascent chain, which allows a favorable substrate geometry for peptide bond formation.