Protein-protein and peptide-protein docking and refinement using ATTRACT in CAPRI
24-Nov-2016
Proteins, DOI: 10.1002/prot.25196
Proteins, online article
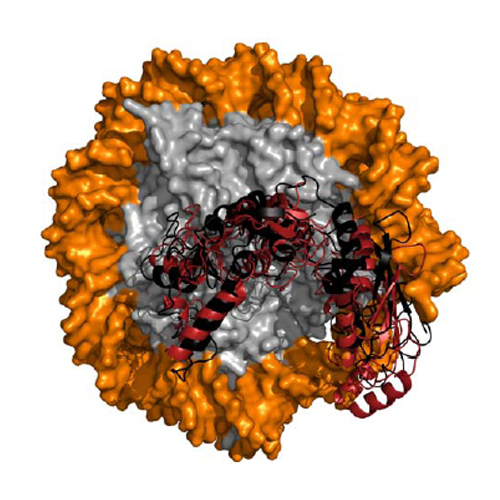
The ATTRACT coarse-grained docking approach in combination with various types of atomistic, flexible refinement methods has been applied to predict protein-protein and peptide-protein complexes in CAPRI rounds 28–36. For a large fraction of CAPRI targets (12 out of 18), at least one model of acceptable or better quality was generated, corresponding to a success rate of 67%. In particular, for several peptide-protein complexes excellent predictions were achieved. In several cases, a combination of template-based modeling and extensive molecular dynamics-based refinement yielded medium and even high quality solutions. In one particularly challenging case, the structure of an ubiquitylation enzyme bound to the nucleosome was correctly predicted as a set of acceptable quality solutions. Based on the experience with the CAPRI targets, new interface refinement approaches and methods for ab-initio peptide-protein docking have been developed. Failures and possible improvements of the docking method with respect to scoring and protein flexibility will also be discussed.