Scientific Strategy
Proteins are the central products of genetic information and are the pivotal biological macromolecules that determine the structure and function of all living systems. Proteins direct organism development, metabolism and responses to environmental stimuli. Traditionally, proteins have been studied as isolated entities. However, in the post-genomic era, the necessity for a new type of protein research has arisen that, whilst continuing investigations on an atomic and molecular level, aims at a more integrated view of protein function within the natural context of living organisms. It is clear today that proteins function mainly in tightly regulated and often transient complexes. This forces basic research to analyze protein function at different levels of complexity using novel and highly sophisticated technologies that are not available in ordinary biochemical and biomedical research labs. Modern NMR techniques allow the study of protein dynamics. Short time spectroscopy is needed to investigate processes such as protein folding and ligand binding. All techniques enable one to study proteins on a molecular level. For the level of protein organization, mass spectrometry is the technique of choice. It not only allows detection of post-translational modifications but also enables one to decipher the components of even transiently formed protein complexes. Single molecule spectroscopy and bioimaging are other tools needed to study proteins on a systems level e.g. in living cells. Access to such cutting edge technologies will allow researchers to capture and analyze molecular protein assemblies in systems with the aim of manipulating complex cellular processes. Modern protein science therefore studies all levels of organization of life, from I) single protein molecules, to II) complexes of proteins with other proteins, ligands, and nucleic acids up to III) the analysis of the highly dynamical protein networks in cells, tissues, and model organisms, such as C. elegans, Drosophila, mouse, zebrafish, Arabidopsis and prokaryotes. Thus, integrated protein science will be a challenging and highly interdisciplinary research endeavour spanning from biophysics to cell biology and genetics. An important aspect of CIPSM will be to provide access to large-scale equipment and funding for new instruments that will be used jointly by several groups.
The overall aim of CIPSM is to foster an integrated protein science approach in order to make seminal contributions to our understanding of protein function in living systems. In addition, we plan to develop new technologies to study, manipulate and quantify protein function in the natural environment. On the fundamental level of biology, we will follow three major lines of research: (I) analysis of protein functions and networks in genome expression and repair, (II) systemic study and intervention of protein function and dysfunction in neurobiology and medicine and (III) analysis of proteins in cellular architectures and for compartmentalisation.
These basic scientific questions will be investigated in six different research areas (A–F), each headed by two experienced coordinators.
Research area A
In research area A biophysics will analyze single proteins using mechanical and optical tools as well as ultrafast spectroscopy on a molecular level. Cutting edge single molecule mechanical techniques such as AFM and optical tweezers will be used to study protein folding (areas B and F), protein nucleic acid interactions (D and partly E) and the function of protein machines (B, C and D). Modern optical single molecule spectroscopy techniques are employed for the analysis of protein folding (areas B and F), protein-protein (area E, Berg) and protein-nucleic acid interactions (area D) as well as protein trafficking in living cells (area E). In addition, time resolved spectroscopy tools (ultrafast IR and UV/Vis) are developed and employed to analyze protein folding/misfolding (areas B and F). The research in the biophysics area will be tightly linked to all other research areas in a network of collaborations with the biologically and biochemistry oriented groups.
Research area B
In research area B, the proteins involved in folding, protein-protein assembly and the machinery behind protein transport in cells will be analyzed using the methods of biochemistry, cell biology and biophysics supplied by the research area A. In area B, analysis of chaperones and the biochemistry behind the coordinated transport of functional proteins inside cells are the major research objectives. ATP driven nano-machines under investigation in area A are playing a fundamental role in this transport process. Area B provides insight that can be directly used by the groups working on protein misfolding in disease states (area B).
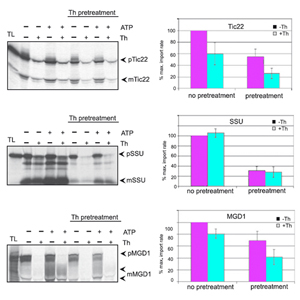
Research area C
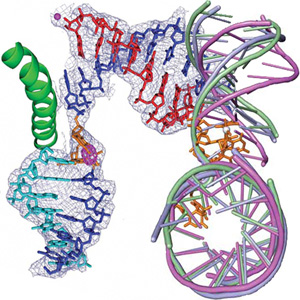
The goals in the research area C are the investigation of the three dimensional structure of proteins including investigation of protein dynamics (by NMR), protein-protein complexes and protein-nucleic acid complexes (in collaboration with groups in area D, E and F). In order to analyse protein-protein complexes, this research area hosts groups from the Max-Planck Institute of Biochemistry, which concentrate at the analysis of protein complexes using mass spectrometry (Mann) and which analyse the rules, which govern the assembly of these complexes (Jentsch). In order to analyze proteins more specifically in cells, the chemists working in area E are developing new chemical proteomics tools. This technique can by combined with the MS approach (MPI, AG Mann). CIPSM has formed a structural biology unit, which includes the Bavarian NMR facility with its 900 MHz NMR spectrometer at the TUM, the structural biology groups from the Gene Center at the LMU and at the WZW as well as cryoelectron microscopy facilities. The groups working in area C interact closely with all groups working on proteins on the atomic and molecular level in CIPSM. For the analysis of protein-nucleic acid interactions, a strong link between this research area C and groups working specifically on protein nucleic acid interactions in the areas D and E was established.
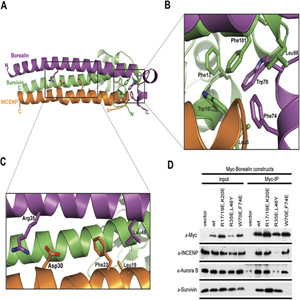
Research area D
The groups working in research area D are specifically looking at protein-DNA and protein-RNA interactions. Chromatin and the whole field of epigenetics, including cellular development are the “buzzwords” in this area. The groups have strong interactions with the structural biology groups in area C as well as with the groups working in chemical biology/genetics (area E). The scientific goal of the collaboration with E is to manipulate the epigenetic programming of cells in order to control development processes. Another goal is to study genome maintenance using damaged DNA substrates directly injected into the cell nucleus or, alternatively, to use chemically modified RNAi tools. Area D is strongly linked to area A by their needs for biophysical tools.
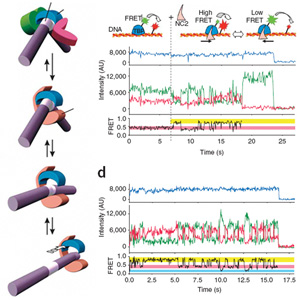
Research area E
In area E we have combined the activities of protein engineering and chemical biology/genetics with the unifying aim to manipulate biomolecular and cellular processes at their causative levels. To this end, proteins with novel functions, created either by rational design, or via evolutionary methods and novel classes of small molecules, which can work as function modulators in living cells will be developed and employed. The desire to funnel protein research into applications is the driving force behind the integration of a strong protein biotechnology section into the CIPSM Cluster. Interesting proteins developed in the other areas can be taken up into the development process. Protein design also involves the methods for structural analysis (area C). In the area E, chemists are preparing new tools to investigate and to measure protein function in living cells (chemical genetics, chemical proteomics). The methods will be used to facilitate the mass spectrometric analysis of functional protein complexes (link to area C). This research area needs the bioimaging facility provided by area A and has extremely intensive links to area D.
Research area F
In research area F CIPSM has bundled together the activities of groups working on disease states. The focus points are neurological diseases associated with protein misfolding. This area will strongly benefit from the research areas A and B which investigate folding problems. CIPSM will strongly profit from groups working directly on health related issues. Strong interactions between the multiphoton spectroscopy used in the area F and the biophysics groups working in area A will boost the technology needed to analyze these processes in vivo. The group of Klein and Carell are currently developing new voltage sensitive fluorescence dyes.
In all six fields of research within CIPSM, multifunctional protein assemblies are of central importance and need to be structurally and functionally analyzed in vitro and in vivo. We therefore plan to strengthen our facilities that allow analysis of protein complexes in vitro, e.g. by structural biology techniques (including NMR), small-molecule approaches and mass spectrometry as well as in vivo genomic screens and cutting edge bio-imaging approaches. CIPSM will provide matching funds to install new cutting edge equipment. This equipment will be instrumental to obtain new mechanistic insights that are expected to enable the manipulation of the molecular processes of gene regulation and neuronal signalling. This in turn will pave the way for new biomedical approaches. In addition, previously unknown proteins will be discovered that may be funnelled into structural analysis and protein engineering in area C and E. On the more applied level of protein science we plan to establish a strong research focus in the fields of protein design and biopharmaceuticals in area E. From the manipulation and detection of proteins with small molecules in living system (area E) we expect a spill over into medicinal applications. This research area will strongly benefit from groups working in the field of protein function and dysfunction in living cells (area F). One can already see how chemists and medicinal groups interact to provide the basics for new therapeutic approaches.
In order to establish even stronger links between the six research areas LMU plans to establish four new full professors. In addition W2 tenure track professorships will be installed to fill further research gaps. TUM has decided to install several strategic W3 and W2 professorships. Several of these are backed up by the InnovaTUM initiative at TUM. These professorships are either an integral part of CIPSM; or will be associated therewith.
On the W2tt level at TUM and LMU, we plan to particularly strengthen research in the fields of I) protein modeling, II) fibrous proteins, III) biophysics, IV) cell architecture. V) hybrid methods in structural biology, VI) developmental biology including stem cell research, VII) peptide biochemistry VIII) cell culture technology and VIIII) neurodegeneration. We will hire new independent tenure-track professors and will better integrate the existing junior groups to foster these areas within CIPSM. The need to include junior groups is particularly evident in research area E. Chemical genetics is a very young research field in Germany, which is not covered in Munich by the presence of full professor groups. We have therefore formed a team of three internationally renowned assistant professors (Berg, Mayer, Meister) working in this area from the MPI of Biochemistry in Martinsried and two teams (Carell, Sieber) in organic chemistry at LMU to represent this timely topic. The junior groups at the MPI for Biochemistry will be supported by CIPSM with personnel and consumable money.
Important research questions in CIPSM that will be addressed spanning the research areas include the following:
-
How do proteins function and how do they dynamically interact with each other and with nucleic acids during inheritance, gene expression, gene regulation, and in networks directing genetics and epigenetics? (All areas)
-
How can classical protein science be combined with comparative genomics, bioinformatics, and systems modeling to achieve a better understanding of biological mechanisms? How can structural biology be combined with the analysis of protein networks into a molecular systems biology approach? (Areas C, D and F) How can we understand protein interactions and specificities on detailed molecular structural level?
-
How can protein engineering/design and chemical biology/genetics be used to answer biological questions on the cellular and organismic level and how to create novel drugs for therapeutic intervention in pathological processes? (Areas D and E)
-
What can be learned about protein behaviour by single-molecule approaches? How can in vivo bioimaging unravel the spatial-temporal regulation of protein networks in response to environmental changes? (Areas A through F)
-
How do defined proteins (e.g. receptors, channels) control the normal function of neural circuits? How are those functional protein networks disturbed during disease, in particular during protein biogenesis and turnover, and protein misfolding, as in the case of Parkinson, Alzheimer, and Huntington pathologies?










